Research Theme
Bioinorganic Chemistry of Metalloproteins Responsible for Signal Transduction
Keywords
Bioinorganic Chemistry, Metalloproteins, Sensor Protein
Transition metal ions and metalloproteins play crucial roles in signal transduction processes in addition to their traditional roles in energy and substance metabolisms. Many responses to metals occur transcriptionally or post-transcriptionally. The metal-responsive transcription factors control the expression of genes encoding proteins responsible for metal homeostasis in cells including metal ions uptake/efflux, intracellular metal trafficking, and biogenesis of metalloproteins. Metal-responsive signal transduction pathways emanating from metal sensing at the cell membrane are also responsible for biological regulation in response to metals. Metal-based sensor proteins are utilized to sense external signals that cannot be sensed by simple sensor proteins without any prosthetic group, in which transition metal ions or metal-containing prosthetic groups act as the active center of signal sensing.
My research interests are foucused on the elucidation of the structural and functional relationships for metal-dependent proteins working in biological signal-transduction systems including metal-based sensor proteins, transition metal ion-sensing transcriptional regulators, and protein machineries responsible for metal ions homeostasis in both prokaryotes and eukaryotes.
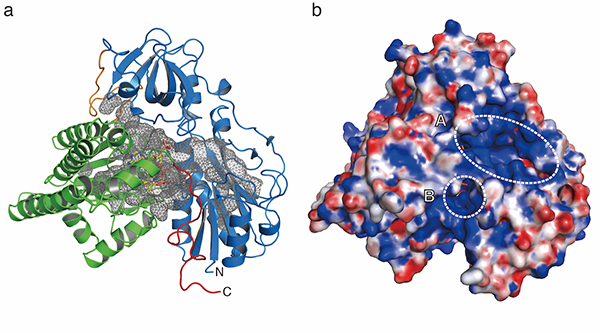
X-ray structure of HypX that catalyzes CO biosynthesis for the assembly of the active site in NiFe-hydrogenase. CoA, which is shown in a stick model, is bound in the cavity (gray mesh in (a)) . There are two open window (A and B in (b)) on the protein surface.
Selected Publications
-
"Heme controls the structural rearrangement of its sensor protein mediating bacterial survival", M. Nishinaga, H. Sugimoto, Y. Nishitani, S. Nagai, S. Nagatoishi, N. Muraki, T. Tosha, K. Tsumoto, S. Aono, Y. Shiro, and H. Sawai, Commun. Biol. 2021, 4, 467 (12 pages) (2021).
-
"Structural characterization of thermoglobin from a hyperthermophilic bacterium Aquifex aeolicus", N. Muraki, K. Takeda, D. Nam, M. Muraki, and S. Aono, Chem. Lett. 2021, 50, 603-606 (2021).
-
"Structural basis for heme transfer reaction in heme uptake machinery from Corynebacteria" N. Muraki, C. K., Y. Okamoto, T. Uchida, K. Ishimori, and S. Aono, Chem. Commun. 55, 13864-13867 (2019).
-
"Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase" N. Muraki, K. Ishii, S. Uchiyama, S. G. Itoh, H. Okumura, and S. Aono, Commun. Biol. 2, 385 (2019).
-
“Protein Dynamics of the Sensor Protein HemAT as Probed by Time-Resolved Step-Scan FTIR Spectroscopy,” A. Pavlou, H. Yoshimura, S. Aono, E. Pinakoulaki, Biophys. J. 114, 584-591 (2018).
-
“Probing the role of the heme distal and proximal environment in ligand dynamics in the signal transducer protein HemAT by time-resolved step-scan FTIR and resonance Raman spectroscopy,” A. Pavlou, A. Loullis, H. Yoshimura, S. Aono, E. Pinakoulaki, Biochemistry, 56, 5309-5317 (2017).