English version 8 October, 2020 on EurekAlert!
Researchers have uncovered details of how a certain type of bacteria breaks down cellulose—a finding that could help reduce the cost and environmental impact of the use of biomass, including biofuel production. The bacteria’s cellulose degradation system is in some way different from how a fungus is already widely used in industry, including to soften up denim to make stone-washed jeans.
-------------------------------------------------------------------
Efforts to find ways to break down cellulose, the tough stuff that makes up plant cell walls, faster and more productively has long been a goal of industrial researchers.
When plants are processed into biofuels or other biomass applications, cellulose has to be degraded into simpler sugar molecules first, and this step can represent up to a quarter of the operating and capital costs of biofuel production. If this process can be made faster and more productive, it won’t just save industry money, but such efficiencies could also reduce the environmental impact of production.
Cellulose molecules bind very strongly to each other, making cellulose very hard to break down. Some fungi are able to break it down, however, and their cellulose degradation systems are well known.
Fungi produce many types of cellulases—enzymes that speed up the chemical reaction that degrades cellulose. And such fungi have been widely used in industry for this reason. For example, the fungus Trichoderma reesei—discovered during World War Two in the Pacific as a result of its eating away at tents and clothing—is used in the production of stone-washed jeans. Cellobiohydrolase, a type of cellulase that the fungus produces, breaks down cellulose into cellobiose, a simple sugar more easily useable by organisms. This slightly degrades the denim material in places, which in turn softens it—making it appear as if washed with stones—and makes it more comfortable to wear.
But there is another type of cellulose degradation system used by some bacteria, and which is similar in many ways to that used by this fungus. But this system has not been very well understood until now. In a paper in the Journal of Biological Chemistry on August 18th, researchers from Japan’s Institute for Molecular Science, National Institutes of Natural Sciences (IMS, NINS) have finally described this system in detail at the single-molecule level.
The type of cellobiohydrolase produced by the bacterium Cellulomonas fimi has a similar catalytic domain to the cellobiohydrolase produced by T. reesei. The catalytic domain of an enzyme is its region that interacts with a molecule that it wants to change or break down (in order to cause the enzymatic reaction). Both the fungus and the bacteria’s cellulose degradation system also exhibit similar hydrolytic activity (the way that they use water to break down the cellulose’s chemical bonds).
But the two systems have different carbohydrate-binding modules (the series of proteins in the enzyme that bind to the carbohydrates in the cellulose) and what are termed “linkers”, in essence the part of the enzyme that links the catalytic domain to the carbohydrate-binding modules.
In earlier research, the NINS scientists had already established that the structure of the linker region of the fungal cellobiohydrolase played a crucial role in how fast the enzyme binds to cellulose (and thus how fast the system degrades cellulose).
“So the obvious next questions were: Even though these other parts of the bacterium’s cellobiohydrolase are different to those of the fungus, do they nevertheless do something similar?” said Akihiko Nakamura and Ryota Iino, the researchers on the team. “Do they also speed up cellulose degradation?”
They found that they do. The scientists used single-molecule fluorescence imaging—an advanced method of microscopy that delivers images of living cells with a resolution of just tens of nanometers—to observe the bacterium’s cellobiohydrolase binding to and dissociating from cellulose molecules.
This allowed them to clarify the functions of the different parts of the cellulose degradation system. They found that the carbohydrate-binding modules were indeed important for the initial binding, but the role played by the linker region was fairly minor.
However, they found that the catalytic domain was not so similar after all. Its structure showed longer loops at the entrance and exit of a “tunnel” in the heart of the system compared to that of the fungus. And this difference in the tunnel structure results in higher processivity—the ability of an enzyme to set off multiple consecutive reactions.
The next steps will be to engineer these bacterial cellulose degrading enzymes to break down cellulose faster.
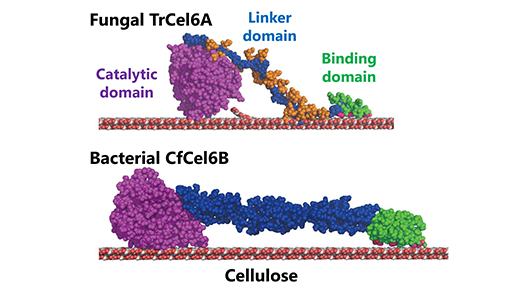
Structural models of fungal and bacterial cellobiohydrolases
(TrCel6A, CfCel6B)
Information of the paper
Authors: Akihiko Nakamura, Daiki Ishiwata, Akasit Visootsat, Taku Uchiyama, Kenji Mizutani, Satoshi Kaneko, Takeshi Murata, Kiyohiko Igarashi, Ryota Iino
Journal Name: Journal of Biological Chemistry
Journal Title: “Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases”
DOI: 10.1074/jbc.RA120.014792
Financial Supports
Grant-in-Aid for Scientific Research on Innovative Areas (JP18H05424、JP18H05425, 18H05494)
Grant-in-Aid for Scientific Research (18H02418、19H03094)
Grant-in-Aid for Research Activity Start-up (15H06898)
Contact Person
Ryota Iino
TEL: +81-564-59-5230
E-mail: iino_at_ims.ac.jp
*Please replace the "_at_" with @
2911

