A team of researchers successfully predicted an unresolved conformation of the oxalate transporter and critical amino acid residues that promote its conformational change by using molecular dynamics simulation and AlphaFold2, the structure prediction AI. This approach, which combines molecular dynamics simulation and structure prediction AI, can be applied to structural studies of various transporter proteins.
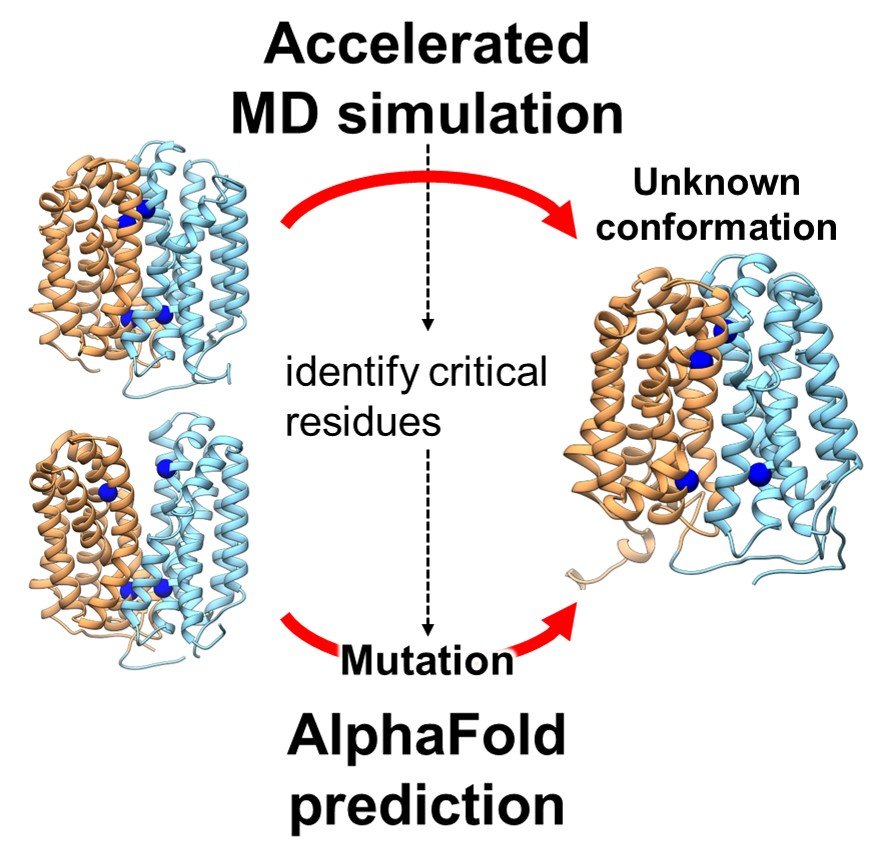
Figure 1:Molecular simulation x AI reveals an unresolved conformation of the oxalate transporter, significant in avoiding kidney stone disease.
In a groundbreaking study, researchers have unveiled a previously unknown conformational state of a crucial transporter protein, OxlT, which plays a vital role in preventing kidney stone formation. This discovery, achieved through advanced computational methods, offers new insights into protein function and potential therapeutic targets.
Proteins are the building blocks of life, performing essential functions in every living organism. Transporter proteins, like OxlT, are particularly important as they carry vital substances across cell membranes. OxlT, found in the oxalate-degrading bacterium Oxalobacter formigenes, is instrumental in managing oxalate levels in the human body. Excess oxalate can lead to kidney stones, a painful and prevalent health issue. Understanding OxlT’s function is crucial, but until now, scientists lacked comprehensive knowledge of its various structural states, particularly the inward-open conformation, a critical part of its transport mechanism.
This study, led by Jun Ohnuki and his colleagues, utilized advanced computational techniques to simulate the OxlT protein’s dynamics. They employed Gaussian accelerated molecular dynamics (GaMD) and AlphaFold2, a cutting-edge machine learning tool, to explore OxlT’s structure and function. The team successfully predicted the elusive inward-open conformation of OxlT, a significant step in understanding its complete functional cycle. This conformation revealed that OxlT prefers binding to formate rather than oxalate in this state, a crucial aspect of its role in oxalate management. Furthermore, the research identified specific amino acid residues critical for this conformational transition, a finding that could have broader implications for understanding protein dynamics.
The implications of this research extend beyond a single protein. The methodology and insights obtained from this study provide a template for exploring other proteins’ dynamics, particularly transporter proteins, which are often targets for therapeutic drugs. Understanding these proteins at a detailed level can lead to the development of more effective treatments for a variety of conditions. Additionally, this research exemplifies the power of combining computational biology with machine learning, a rapidly evolving field that promises to unlock many of biology’s most challenging mysteries. By filling a crucial gap in our understanding of the OxlT protein, this study not only contributes to potential advancements in kidney stone prevention but also paves the way for future breakthroughs in biomedical research.
The research team includes Jun Ohnuki, Titouan Jaunet-Lahary and Kei-ichi Okazaki from the Research Center for Computational Science at Institute for Molecular Science (IMS), NINS. Completing the team is Atsuko Yamashita from the Graduate School of Medicine, Dentistry and Pharmaceutical Sciences at Okayama University.
Information of the paper:
Authors: Jun Ohnuki, Titouan Jaunet-Lahary, Atsuko Yamashita, and Kei-ichi Okazaki
Journal Name: The Journal of Physical Chemistry Letters
Journal Title: “Accelerated Molecular Dynamics and AlphaFold Uncover a Missing Conformational State of Transporter Protein OxlT”
DOI: 10.1021/acs.jpclett.3c03052
Financial Supports:
KAKENHI, Grant-in-Aid for Early-Career Scientists: JP23K14160
KAKENHI, Grant-in-Aid for Scientific Research(B): JP22H02595
Nagase Science and Technology Foundation
Research Center for Computational Science: 22-IMS-C189,23-IMS-C201
Contact Person:
Name: Kei-ichi Okazaki
Research Center for Computational Science, Institute for Molecular Science, NINS
The Graduate University for Advanced Studies, SOKENDAI
TEL: +81-564-55-7468
E-mail: keokazaki_at_ims.ac.jp (Please replace the "_at_" with @)
3467

