Computer simulations revealed the detailed mechanism of how the protein "dynamin" works to form small vesicles within cells.
While dynamin uses GTP hydrolysis energy to change shape, it was unclear how this leads to membrane constriction. Simulations showed that instead of simply tightening, dynamin "loosens" (expands) at a certain stage to generate the force needed to narrow the surrounding membrane tube.
This study provides a clearer explanation for membrane deformation and vesicle formation processes in cells, offering insights for artificial nano-device design.
Researchers have used sophisticated coarse-grained molecular dynamics (CG-MD) simulations to resolve the fundamental mechanism of how the protein dynamin constricts cellular membranes during endocytosis. Contrary to the common notion that dynamin directly tightens around the membrane like a belt, the simulations demonstrated that the hydrolysis of guanosine triphosphate (GTP) causes the dynamin ring to "loosen" and expand. This ring expansion results in an unexpected and crucial indirect constriction of the adjacent membrane region.
Dynamin is a protein that plays a central role in endocytosis--the process where cells internalize substances by wrapping them in cell membrane vesicles. For a vesicle to detach, the neck of the membrane must be constricted and cut. Dynamin assembles into a ring shape around this neck and uses the energy from hydrolyzing GTP to change its shape and sever the membrane (Figure 1). However, observing exactly how this structural change--specifically the transition from the GTP-bound state to the GDP-bound state--mechanically leads to membrane constriction has been difficult experimentally due to the protein's large and complex structure.
To investigate this molecular mechanism, the research team employed coarse-grained molecular dynamics (CG-MD) simulations. This computational approach allowed them to construct a realistic system containing both the large dynamin assembly and the lipid membrane tube (Figure 1). By simulating the transition from the GTP state to the GDP state, they successfully reproduced the detailed movements of dynamin and the membrane.
The simulation results provided a new perspective: dynamin does not simply "squeeze" the membrane. Instead, upon GTP hydrolysis, the dynamin ring structure "loosens" or expands. This expansion pushes against the membrane in the coated area, which mechanically induces a significant indirect constriction in the adjacent, protein-uncoated region of the membrane tube (Figures 1 and 2). This force narrows the membrane tube sufficiently to facilitate fission.
By successfully reproducing the complex interplay between protein structural changes and membrane deformation on a computer, this research significantly advances our understanding of various biological phenomena, including intracellular transport and viral entry. Furthermore, these findings regarding the mechanics of membrane deformation pave the way for engineering applications, such as the design of artificial nano-devices, and could contribute to medical research concerning diseases caused by malfunctions in membrane dynamics.
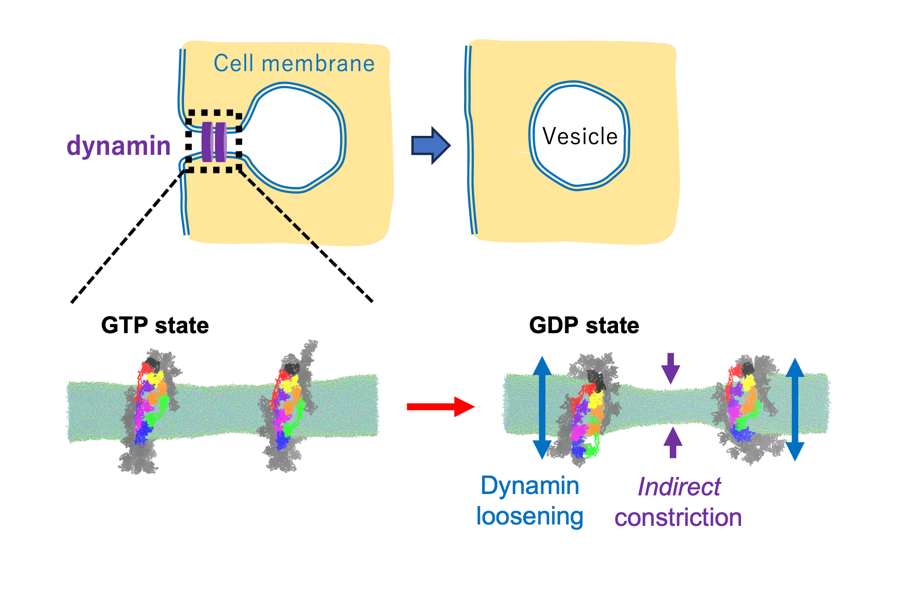
Figure 1: Dynamin ring constricts the membrane by "loosening."
Dynamin assembles into a ring around the neck of a forming vesicle and utilizes GTP hydrolysis energy to constrict and sever the membrane. The simulation reveals that the dynamin ring expands (loosens) upon GTP hydrolysis, which indirectly forces the adjacent membrane tube to narrow. (Credit: Kei-ichi Okazaki, Restriction: CC BY)

Figure 2: Simulations of indirect membrane constriction by dynamin.
The simulation reveals that the dynamin ring expands (loosens) upon GTP hydrolysis, which indirectly forces the adjacent membrane tube to narrow. (Credit: Kei-ichi Okazaki, Restriction: CC BY)
Information of the paper
Authors: Md. Iqbal Mahmood, Shintaroh Kubo, Hiroshi Noguchi, and Kei-ichi Okazaki
Journal Name: The Journal of Physical Chemistry Letters
Journal Title: "Membrane Constriction by Dynamin through GTP-Driven Conformational Changes from Coarse-Grained Molecular Dynamics Simulations"
DOI: 10.1021/acs.jpclett.5c02867
Financial Supports
Grant-in-Aid for Scientific Research(B): JP22H02595, JP23K23858
Grant-in-Aid for Transformative Research Areas (A) : JP25H02299
Grant-in-Aid for Early-Career Scientists: JP22K15070
Grant-in-Aid for Scientific Research (C): JP24K06973
Contact Person
Kei-ichi Okazaki
Research Center for Computational Science, Institute for Molecular Science, NINS
TEL: +81-564-55-7468
E-mail: keokazaki_at_ims.ac.jp (Please replace the "_at_" with @)
3467


