Research Theme
Dynamical Ordering of Biomolecular Systems for Creation of Integrated Functions
Keywords
Biomolecule, Dynamical Ordering, NMR
Living systems are characterized as dynamic processes of assembly and disassembly of various biomolecules that are self-organized, interacting with the external environment. The omics-based approaches developed in recent decades have provided comprehensive information regarding biomolecules as parts of living organisms. However, fundamental questions still remain unsolved as to how these biomolecules are ordered autonomously to form flexible and robust systems. Biomolecules with complicated, flexible structures are selforganized through weak interactions giving rise to supramolecular complexes that adopt their own dynamic, asymmetric architectures. These processes are coupled with expression of integrated functions in the biomolecular systems.
Toward an integrative understanding of the principles behind the biomolecular ordering processes, we conduct multidisciplinary approaches based on detailed analyses of dynamic structures and interactions of biomolecules at atomic level, in conjunction with the methodologies of molecular and cellular biology along with synthetic and computational technique.
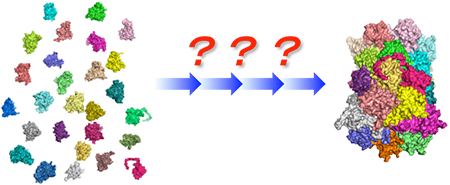
Formation of supramolecular machinery through dynamic assembly and disassembly of biomolecules.
Selected Publications
- K.Kato, S.Yanaka, and T.Yamaguchi, “The synergy of experimental and computational approaches for visualizing glycoprotein dynamics: Exploring order within the apparent disorder of glycan conformational ensembles”, Curr. Opin. Struct. Biol. 92, 103049 (2025)
- K.Kato, S.Yanaka, and H.Yagi, “Technical basis for nuclear magnetic resonance approach for glycoproteins”, Experimental Approaches of NMR Spectroscopy II (The Nuclear Magnetic Resonance Society of Japan ed.), Springer Nature Singapore, pp.169-195 (2025)
- H.Yagi, K.Takagi, and K.Kato, “Exploring domain architectures of human glycosyltransferases: Highlighting the functional diversity of non-catalytic add-on domains,” Biochim. Biophys. Acta –General Subjects, 1868, 130687 (2024)
- D. Koga, S.Kusumi, H.Yagi, and K.Kato, “Three-dimensional analysis of the intracellular architecture by scanning electron microscopy”, Microscopy, 73, 215-225 (2024)
- K.Kato and H.Yagi, “Current status and challenges in structural glycobiology,” Trends in Carbohydrate Research, 15, 38-46 (2023).
- Yagi-Utsumi, M. and Kato, K., “Conformational variability of amyloid-β and the morphological diversity of its aggregates,” Molecules 27, 4787 (2022).