Research Theme
Elucidation and Control of Dynamics of Biomolecular Machines in Function
Keywords
Biomolecular Machines, Theoretical Biophysics, Molecular Simulations
Biomolecular machines, such as molecular motors and transporters working in the cell, are known to change their structure when they function. For example, ATP synthase, which synthesizes ATP in mitochondria, is a molecular motor that uses chemical energy to rotate. Transporters, which transport substrate molecules across the cell membrane, perform substrate transport by changing their structure between an inwardly and outwardly open structure relative to the membrane. Our goal is to elucidate the mechanism of these elaborate and dynamic nanomachines created by nature at the atomic and molecular level, and to control their functions based on our findings.
We would like to understand the mechanism of biomolecular machines by “seeing” the motion of biomolecular machines at the moment they function, on a computer at the atomic and molecular level. However, this is not an easy task, because biomolecular machines are huge molecules, and their functioning time scale is slow (for a molecular scale) at milliseconds or more. It is difficult to simulate the millisecond time scale motion of a huge system consisting of hundreds of thousands of atoms or more using conventional methods. Therefore, we are trying to capture the motion at the moment of function by using methods such as an importance sampling technique, or coarse-graining multiple atoms together.
We have been working on ATP synthase, which is the main energy source in the cell 1), and Na+/H+ antiporter 2,3), which regulates ion concentrations in the cell. For the Na+/H+ antiporter, we captured the moment of ion transport by simulations and identified the key interactions that regulate ion transport. By experimentally modulating this interaction, we succeeded in increasing the ion transport rate more than twofold. In this way, we will challenge functional control based on the mechanism of the biomolecular machine elucidated by simulation.
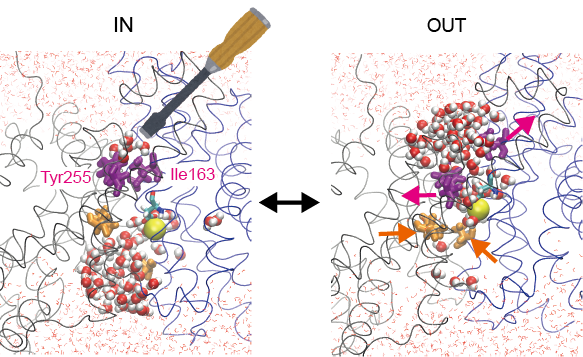
Control of the transporter function based on mechanism elucidated by simulation.
Selected Publications
-
K. Okazaki & G. Hummer, “Elasticity, friction, and pathway of gamma-subunit rotation in FoF1-ATP synthase.” Proc. Natl. Acad. Sci. USA 112:10720-10725 (2015).
-
K. Okazaki, D. Wöhlert, J. Warnau, H. Jung, Ö. Yildiz, W. Kühlbrandt and G. Hummer “Mechanism of the electroneutral sodium/proton antiporter PaNhaP from transition-path shooting”, Nature Communications 10, 1742 (10 pages) (2019).