Research Theme
Biomolecular Dynamics Simulation: Protein Folding, Denaturation, Aggregation, and Amyloid Fibril
Keywords
Molecular Dynamics Simulation, Protein, Amyloid
Biomolecules such as proteins and peptides have complicated free-energy landscapes with many local minima. Conventional molecular dynamics (MD) simulations easily get trapped in a few local-minimum states. We have proposed new generalized-ensemble algorithms, such as the replica-permutation method, to overcome these difficulties. We applied these methods to several proteins and peptides and succeeded in finding the native structures (Fig. 1).
We are also interested in its application in medicine. There is a disease called misfolding disease, which is caused by misfolded proteins. Alzheimer's disease and Huntington's disease are examples. These diseases are caused by misfolded proteins that form spherical aggregates called oligomers or needle-like aggregates called amyloids. However, the mechanism by which these aggregates are formed is still poorly understood. We have identified the amino acids important for forming Alzheimer's disease-causing aggregates by replica-permutation MD simulations. We have also elucidated how these aggregates can be destroyed by ultrasound or infrared laser by non-equilibrium MD simulations (Fig. 2).
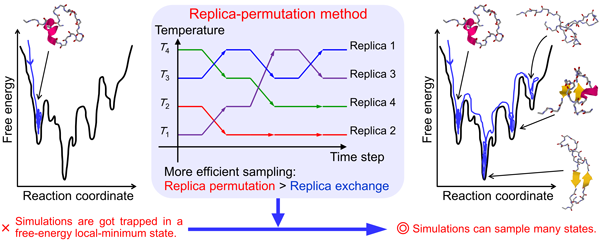
Fig. 1 Biomolecular systems have many free-energy local minima. Conventional molecular dynamics methods are trapped by these minima. We solved this problem by using the replica-permutation method.
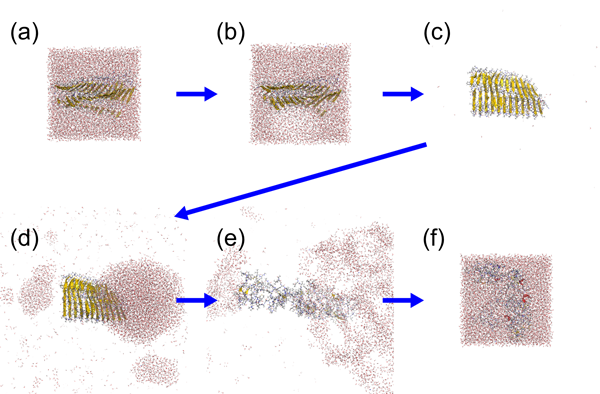
Fig.2 Disruption process of an amyloid fibril of Aβ peptides by ultrasonic wave. The amyloid fibril is disrupted when a bubble collapses.
Selected Publications
-
S. G. Itoh, M. Yagi-Utsumi, K. Kato, and H. Okumura: “Key residue for aggregation of amyloid-β peptides”, ACS Chem. Neurosci. 13, 3139–3151 (2022).
-
H. Okumura, S. G. Itoh, K. Nakamura, and T. Kawasaki: “Role of water molecules in the laser-induced disruption of amyloid fibrils observed by nonequilibrium molecular dynamics simulations”, J. Phys. Chem. B 125, 4964–4976 (2021).
-
H. Okumura and S. G. Itoh, “Amyloid fibril disruption by ultrasonic cavitation: Nonequilibrium molecular dynamics simulations,” J. Am. Chem. Soc. 136, 10549–10552 (2014).
-
S. G. Itoh and H. Okumura: “Replica-permutation method with the Suwa-Todo algorithm beyond the replica-exchange method”, J. Chem. Theory Comput. 9, 570–581 (2013).