A research team headed by Prof. Shigetoshi Aono of the Okazaki Institute for Integrative Bioscience and the Institute for Molecular Science, National Institutes of Natural Sciences, and Prof. Yoshitsugu Shiro, at RIKEN SPring-8 Center, RIKEN Harima Institute, identified a protein that regulates the intracellular concentration of heme1, an iron-containing compound that is essential for life. The researchers revealed, for the first time, the heme regulatory mechanism at the atomic level.
1. Background
By making a complex with a protein, the iron-containing compound heme plays a vital role in various physiological processes in the human body, including the transport and storage of oxygen. But free heme—heme that has not been taken up by proteins—is highly toxic to cells because it generates reactive oxygen species2. For protection against free heme toxicity, many organisms are equipped with a system to precisely regulate the intracellular concentration of heme. Prior to this research, however, the underlying mechanism was not known in any organism.
2. Results
Using biochemical and X-ray crystallographic techniques3, our research team successfully identified a protein that senses toxic free heme and triggers the excretion of heme from cells. We then elucidated the mechanism for maintaining a certain intracellular concentration of heme.
To uncover the mechanism of heme regulation, we used a lactic acid bacterium, which is used to produce fermented dairy product such as cheese, as a model organism. Although not capable of heme biosynthesis, this bacterium takes up heme its surroundings for heme-dependent physiological functions. We successfully identified a heme-sensing protein that functions as a molecular switch for the heme regulatory mechanism in this bacterium (Fig. 1). When heme is not present in the cell, this protein binds to a gene responsible for exporting heme (heme-exporter gene) and inhibits the expression of the gene. When the intracellular concentration of heme becomes high, this heme-sensing protein stops binding to the heme-exporter gene and makes a complex with heme, thus turning on the gene expression and the heme export system.
To clarify how this heme-sensing protein functions as a molecular switch for the heme export system, we performed X-ray crystallography at SPring-84, the world’s largest synchrotron radiation facility, and we investigated the structure of the protein in its three different forms: with heme, without heme, and with the heme-exporter gene without heme. We determined the position of each atom in the three-dimensional structures and found that, to make a complex with heme, two of the histidine residues in the amino acid sequence form bonds with a heme molecule (His72 and His149 in Fig. 2). When the protein is not making a complex with heme, the peptide structure near His72 appears like a string of atoms. Upon binding to heme, however, this structure adopts a helix conformation (Fig. 2, orange part). Due to this conformational change, the DNA-binding domain of the protein that interacts with the heme-exporter gene (Fig. 1, green part) is bent upward, making it unable to interact with the gene. These findings revealed, at the atomic level, how the protein senses free heme and switches on the heme export system through the conformational change.
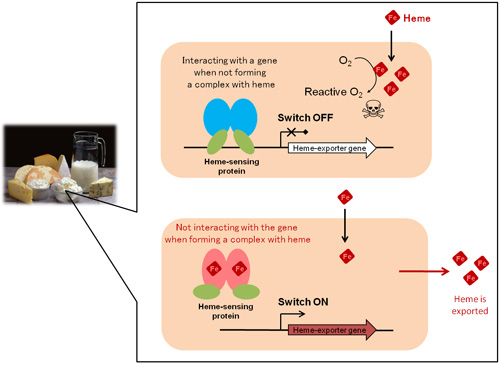
Figure 1. The mechanism of regulating intracellular heme concentrations in lactic acid bacterium Cream-colored boxes represent bacterial cells.
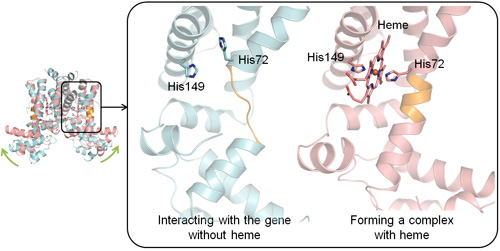
Figure 2. Differences in the three-dimensional structures with and without heme
The illustration on the left shows overlapping images of the two conformations of protein under different conditions: blue, interacting with the heme-exporter gene; pink, making a complex with heme. The right panel shows enlarged images of the domain where the conformational change occurs. His denotes histidine.
3. Social significance of this study
Although we investigated the heme export system of a beneficial lactic acid bacterium in this study, some disease-causing bacteria have similar systems. Therefore, the results of this study can be used to develop new types of antibiotics that inhibit the growth of disease-causing bacteria by making them unable to avoid free heme toxicity. Because we succeeded in clarifying the positions of atoms in the heme-sensing protein by using X-ray crystallographic techniques, highly effective pharmaceutical products can now be designed that are capable of regulating heme-related mechanisms in cells with minimal adverse effects.
4. Glossary
1) Heme
A compound containing an iron atom in the center of the planar cyclic ring called porphyrin. Porphyrins are classified into several groups depending on the type and place of ring modification. Hemoproteins are defined as proteins that are functional only after incorporating a heme molecule, and in general, they appear red. Representatives of hemoproteins are hemoglobin, which delivers oxygen, cytochromes in the electron transport system, and peroxidases, which functions as an enzyme.
2) Reactive oxygen species
A collective term for chemical species containing highly reactive oxygen. Some examples are OH• (hydroxyl radical), O2•− (superoxide anion radical), and H2O2 (hydrogen peroxide). These chemicals cause oxidative damage to biological molecules, such as DNA, lipids, and proteins, resulting in the loss of the normal physiological functions of these molecules.
3) X-ray crystallography
A technique used to determine the structures of molecules. This is achieved by using a beam of short-wavelength X-rays to strike a crystal with an orderly arrangement of atoms and carefully analyzing the intensity of the scattered X-rays. The three-dimensional structures of a variety of biological molecules have been determined by this method.
4) The world’s largest synchrotron radiation facility, SPring-8
A research facility located in Harima Science Park City in Hyogo Prefecture. This facility is managed by RIKEN and is capable of delivering what is now the strongest radiation in the world. At SPring-8 which stands for Super Photon ring-8 GeV (= 8 billion electron volts), waves of electromagnetic radiation are generated by accelerating electrons to near the speed of light and then bending the direction of electrons using a magnetic field. Electromagnetic radiation generated in this way is called synchrotron radiation, which has been widely used for research in a wide range of disciplines and industries, including nanotechnology and biotechnology.
5. Information of the paper
Journal:Journal of Biological Chemistry (J. Biol. Chem.)
DOI: 10.1074/jbc.M112.370916
Title of the paper:Structural basis for the transcriptional regulation of heme homeostasis in Lactococcus lactis
Authors:Hitomi Sawai, Masaru Yamanaka, Hiroshi Sugimoto, Yoshitsugu Shiro, Shigetoshi Aono
Date:July 13, 2012 (e-Pub ahead of print)
6. Research group
This work was performed by collaboration between Drs. Shigetoshi Aono (professor), Hitomi Sawai (research assistant professor), Masaru Yamanaka (postdoctral researcher) of the Okazaki Institute for Integrative Bioscience and the Institute for Molecular Science, National Institutes of Natural Sciences and Drs. Yoshitsugu Shiro (chief scientist) and Hiroshi Sugimoto (senior research scientist) at RIKEN/SPring-8 Center.
7. Financial supports
This work was partly supported by Grant-in-Aid for Scientific Research B (23350084) and Grant-in-Aid for Research Activity Start-up (22870043) from the Japan Society for the Promotion of Science, by the Noda Institute for Scientific Research, by the Funi Yamamura Memorial Foundation for Female Natural Scientists from Chuo Mitsui Trust and Banking, and by the SHISEIDO Female Researcher Science Grant from Shiseido Co., Ltd.
8. Contact
Shigetoshi Aono
Professor, Institute for Molecular Science
TEL: +81-564-59-5575 FAX: +81-564-59-5576 E-mail:aono@ims.ac.jp
Home page: http://www.ims.ac.jp/english/know_en/bio/aono/aono_en.html
1119


